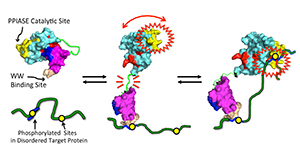
 NMR-based model for Pin1, and its interaction with its “disordered” protein targets pertinent to cancer and Alzheimer’s disease (green). (1) Close contact between “A” and “B” acts as a “brake” on “A” activity. (2) Upon target encounter, “B” binds one site, which unleashes “A,” thereby releasing the brake on “A” activity. (3) The flexibly tethered “A” is then poised to hunt for an adjacent site with enhanced activity.
NMR-based model for Pin1, and its interaction with its “disordered” protein targets pertinent to cancer and Alzheimer’s disease (green). (1) Close contact between “A” and “B” acts as a “brake” on “A” activity. (2) Upon target encounter, “B” binds one site, which unleashes “A,” thereby releasing the brake on “A” activity. (3) The flexibly tethered “A” is then poised to hunt for an adjacent site with enhanced activity.
Drugs to treat cancer and Alzheimer’s disease usually target the active sites of specific protein molecules sustaining the disease. Traditional drug design views proteins as rigid 3-D objects with active sites consisting of surface-accessible “pockets” with a specific, well-defined structure. Traditional drug design involves finding small molecules with shapes that fit specifically into this pocket. A new study from University of Notre Dame researchers suggests that there are alternative approaches to targeting these proteins, a significant finding for future clinical applications.
“Proteins are the nanometer-sized ‘devices’ encoded by our DNA,” Jeffrey Peng, associate professor of chemistry and biochemistry and lead author of the study, said. Depending on their sequences, Peng said, proteins fold into various three-dimensional shapes capable of carrying out distinct functions. “These functions often involve binding to another protein molecule to turn on/off crucial cell signals, such as those controlling the cell cycle. Normally those interactions are under strict regulation. Deregulation of these interactions can lead to hyperactive signaling. If these signals impact the cell cycle, the end result can be cancer or Alzheimer’s disease.”
This realization is a key incentive for the design of drug molecules that bind specific proteins, thereby blocking or inhibiting their natural interactions, which may be in overdrive. The traditional drug design process typically assumes the protein shapes are static. But tradition turns out to be too limiting.
“Signaling proteins, such as those pertinent to cancer and Alzheimer’s disease, have dynamic architectures and are quite flexible,” Peng said. “These proteins’ shapes can morph upon binding. Predicting which parts of the protein are crucial for specific binding becomes significantly more challenging.”
Understanding how these more flexible proteins bind their targets challenges both basic biophysics and targeted drug therapy. Peng has been addressing this challenge through studies of Pin1, a flexible signaling protein important for both cancer and Alzheimer’s disease research.
“There’s recent evidence suggesting that people more susceptible to certain cancers are less likely to develop Alzheimer’s disease, and vice versa,” Peng said. “This makes sense given that both diseases depend on signals driven by Pin1 interacting with other proteins.”
Pin1 consists of two flexibly linked globular units or domains.
“It’s sort of like a two-ball bolas, with the two balls corresponding to two domains that perform distinct yet complementary functions,” Peng said.
 Jeffrey Peng
Jeffrey Peng
Previous studies indicated that Pin1’s two domains communicate with each other in order to bind its targets. But how?
“What’s been missing — and what is essential for drug design — is an atomic-level explanation for long-range communication between the two domains,” Peng said.
A related mystery has been how Pin1 binds its target proteins, which typically have multiple Pin1 binding sites within highly dynamic “intrinsically disordered regions.” These disordered regions challenge the “structure determines function” orthodoxy. Yet they are increasingly recognized as functional units, figuring prominently in cancer-related proteins. So a second question has been, how does Pin1 interact with those disordered sites?
Using high-field nuclear magnetic resonance (NMR) spectroscopy, Peng and his group propose answers to both questions. He explained with a metaphor.
“When Pin 1 is by itself, its two domains, call them ‘A’ and ‘B,’ have greater physical contact,” he said. “The enhanced contact provides ‘braking action’ on the ‘A’ domain catalytic activity. But when Pin1 encounters its protein domain target, the ‘B’ domain reduces its contact with the ‘A’ domain to grab onto the target. This releases the brake (interdomain contact), causing increased activity of the ‘A’ domain.
“This long-range braking action enables dynamic communication between Pin1’s domains. And it could explain how Pin1 is able to interact with multiple docking sites in those disordered protein regions. Pin1 could bind to the first docking site via its ‘B’ domain, tethering Pin1 as a whole, and also reducing interdomain contact, and turning the ‘A’ domain to ‘full power.’ The “A’ domain, still flexibly tethered to the bound ‘B’ domain, could then ‘hunt’ for another distinct docking site, and carry out its function, which is to accelerate local structural changes in the target.
The finding has significant future clinical implications.
“Drugs usually directly target the active sites of proteins,” Peng said. “Typically, they are inhibitors that block binding of the original target. But our results show that sites outside of the active site — namely, the sites of interdomain contact — also affect the Pin1 activity by changing its domain dynamics. So, these allosteric sites site could instead be targeted, as opposed to the active site itself. We show new avenues of inhibition, such as in cancer therapy aimed at inhibiting hyperactive Pin1 in cancer cells.”
Co-authors of the study are Xingsheng Wang, research scientist in the Department of Chemistry and Biochemistry; Brendan J. Mahoney and Meiling Zhang, graduate students in chemistry and biochemistry; and John S. Zintsmaster, Department of Chemistry and Biochemistry. The study appears in the journal Structure and can be found here: www.cell.com/structure/abstract/S0969-2126%2815%2900380-9.
Contact: Jeffrey Peng, 574-631-2983, jpeng@nd.edu